Abstract
Human eyes can recognize person identities based on some small salient regions. However, such valuable salient information is often hidden when computing similarities of images with existing approaches. Moreover, many existing approaches learn discriminative features and handle drastic viewpoint change in a supervised way and require labeling new training data for a different pair of camera views. In this paper, we propose a novel perspective for person re-identification based on unsupervised salience learning. Distinctive features are extracted without requiring identity labels in the training procedure. First, we apply adjacency constrained patch matching to build dense correspondence between image pairs, which shows effectiveness in handling misalignment caused by large viewpoint and pose variations. Second, we learn human salience in an unsupervised manner. To improve the performance of person re-identification, human salience is incorporated in patch matching to find reliable and discriminative matched patches. The effectiveness of our approach is validated on the widely used VIPeR dataset and ETHZ dataset.
Contribution Highlights
- An unsupervised framework is proposed to extract distinctive features for person re-identification without requiring manually labeled person identities in the training procedure.
- Patch matching is utilized with adjacency constraint for handling the misalignment problem caused by viewpoint change, pose variation and articulation. We show that the constrained patch matching greatly improves person re-identification accuracy because of its flexibility in handling large viewpoint change.
- Human salience is learned in an unsupervised way. Different from general image salience detection methods, our salience is especially designed for human matching, and has the following properties. 1) It is robust to viewpoint change, pose variation and articulation. 2) Distinct patches are considered as salient only when they are matched and distinct in both camera views. 3) Human salience itself is a useful descriptor for pedestrian matching.
Reference
If you use our codes or dataset, please cite the following papers:
Images
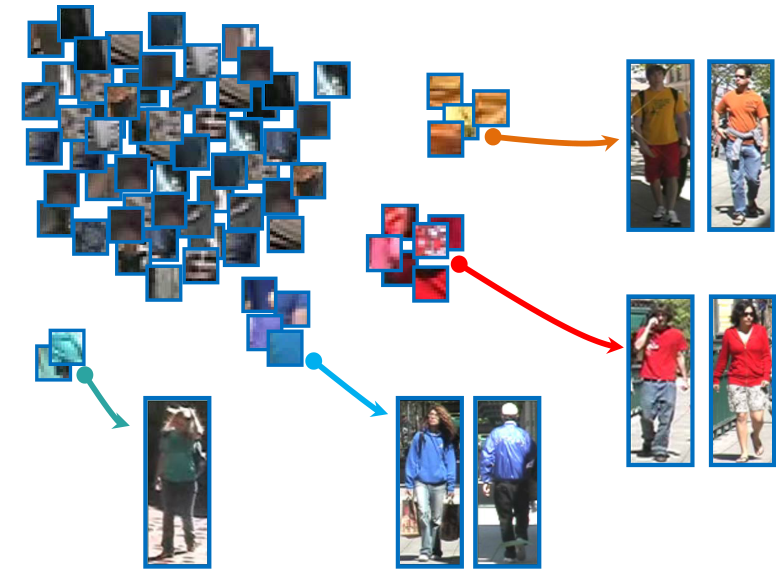
Local patches are densely sampled, and five exemplars are shown in red boxes (within the image). Top ten nearest neighbor patches are shown in the right. Note that the ten nearest neighbors are from ten different images.
Salient patches are those distributed far way from other pathes. We assume that if a person has such unique appearance, more than half of the people in the reference set are dissimilar with him/her.
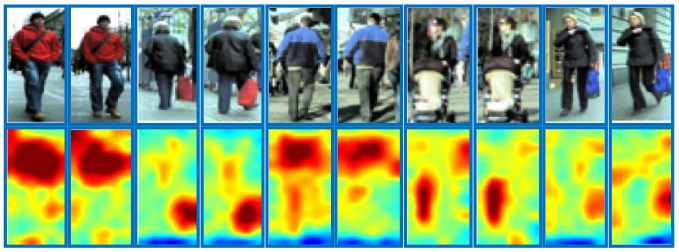
Patches in red boxes are matched in dense correspondence with the guidence of corresponding salience scores in dark blue boxes.
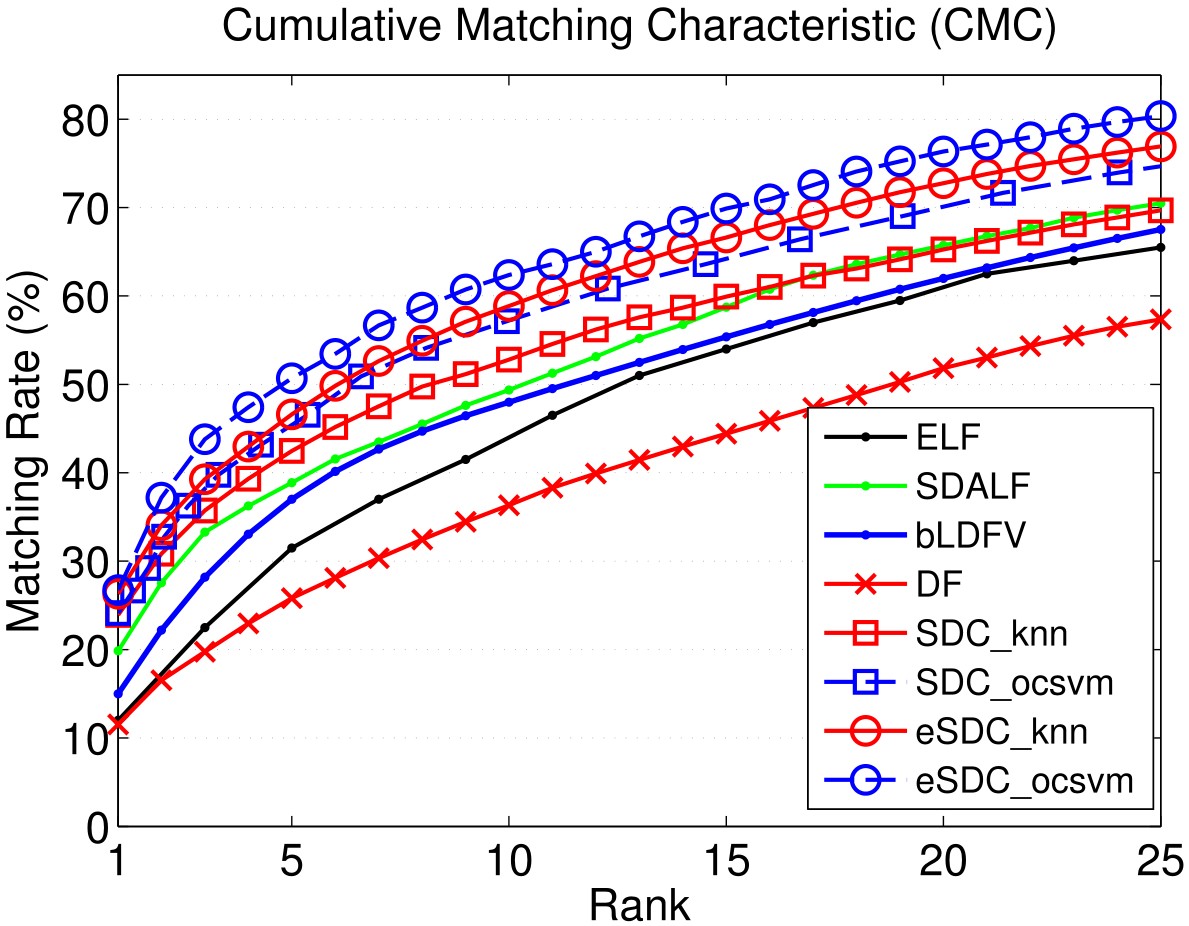
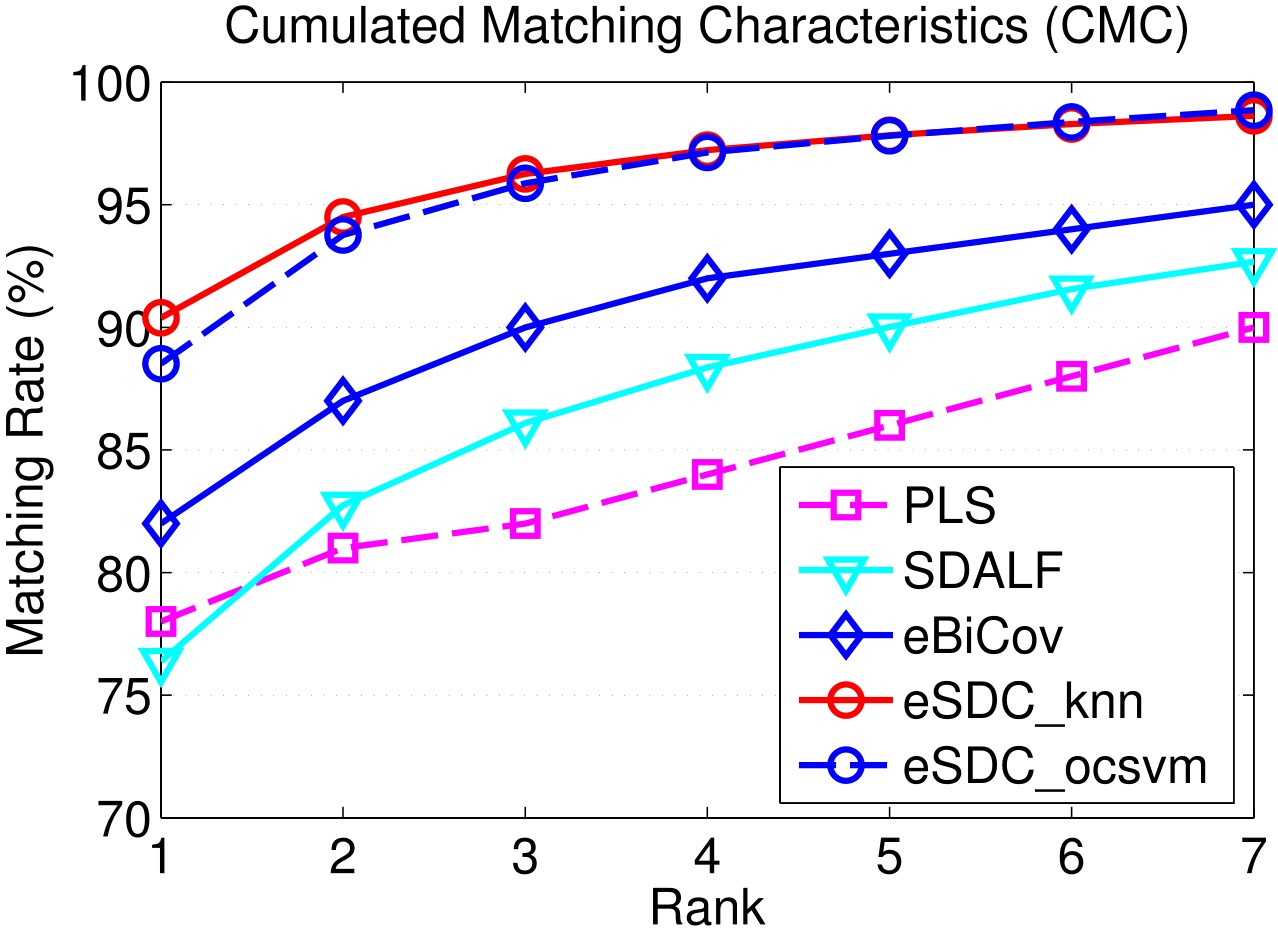
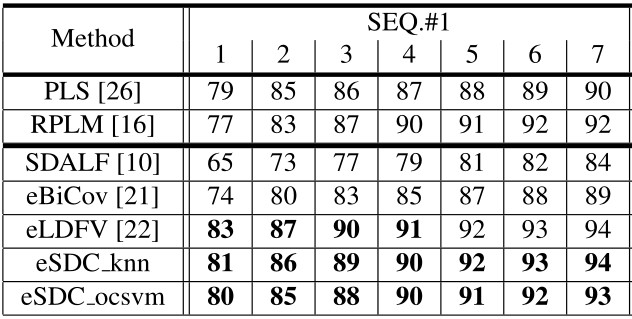
Our approach: SDC knn and SDC ocsvm. Our approach combined with wHSV and MSCR: eSDC knn and eSDC ocsvm.
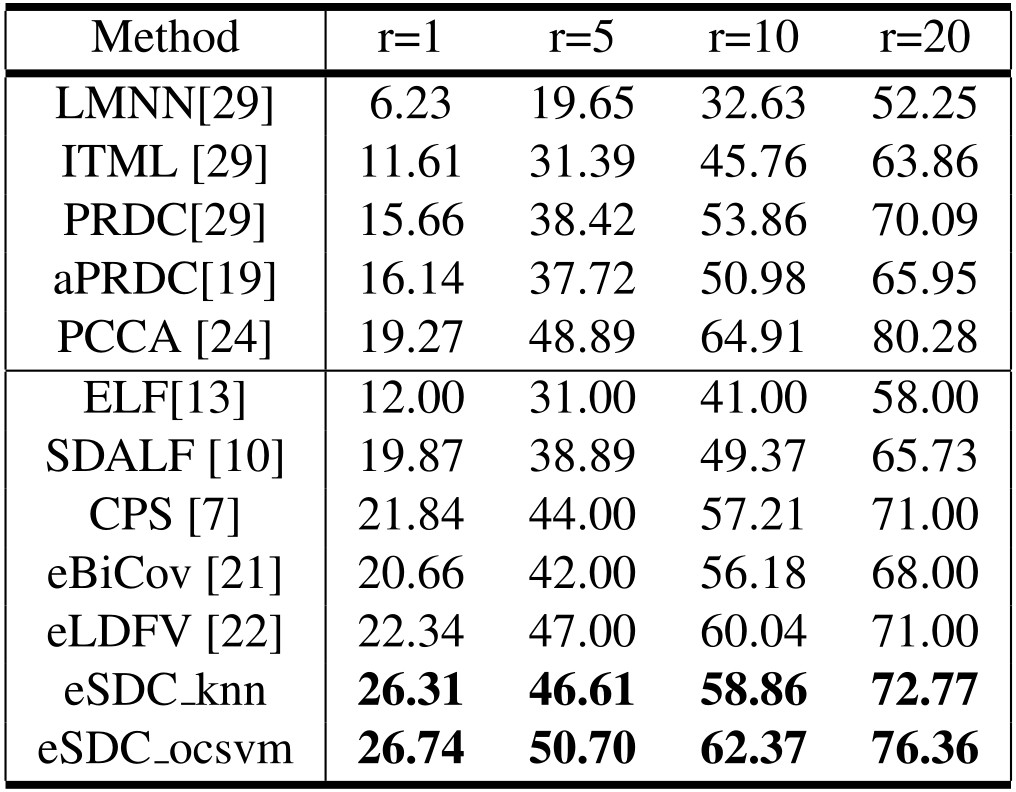
Our approach: SDC knn and SDC ocsvm. Our approach combined with wHSV and MSCR: eSDC knn and eSDC ocsvm. Methods in upper part of the table are supervised while those in lower part are unsupervised methods.
Results include CMC curves on SEQ.#1, SEQ.#2, and SEQ.#3 of the ETHZ dataset. By convention, only the first 7 ranks are shown. All the compared methods are reported under single-shot setting.
Codes
- Dense ColorSIFT feature
- Salience for Re-identification